Chemical synthesis of DNA
Chemical synthesis of DNA
At present, artificial fragments of nucleic acids of different lengths and any composition can be synthesized quite quickly (such fragments are called oligonucleotides), and then combine them into longer chains using special enzymes. The genes and their fragments obtained in this way are widely used in genetic engineering, biotechnology, and also for the diagnosis of infectious and genetic diseases. So chemically synthesized oligonucleotides can be used:
For the construction of whole genes or their fragments
For amplification of specific DNA fragments
For directed mutations of isolated DNA
As probes for hybridization
As linkers to facilitate cloning.
Recall that nucleic acids are irregular biopolymers consisting of monomeric structures - nucleotides (Fig. 29).
Rice. 29. Composition and structure of the nucleotide.
Thus:
Nucleotide \u003d nucleoside + phosphoric acid \u003d nitrogenous base + pentose + phosphoric acid.
In RNA, pentose is ribose. DNA contains deoxyribose.
The composition of nucleic acids and oligonucleotides includes five nucleotides that differ from each other in the structure of the nitrogenous heterocycle. Three of them - derivatives of adenine (A), guanine (G) and cytosine (C) are included in both DNA and RNA, thymine (T) - only in DNA, uracil (U) - only in RNA. Nucleotides, unlike nucleosides, contain a phosphoric acid residue. Nucleotides are connected to each other in a polymer chain using phosphodiester bonds (Fig. 30). Nitrogenous bases do not take part in the connection of nucleotides of one chain.
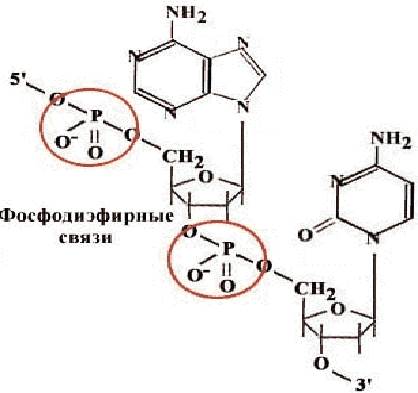
Rice. 30. Scheme of connecting nucleotides using a phosphodiester bond
The task of the chemical synthesis of nucleic acids is the combination of their constituent monomers in a strictly defined sequence. For the formation of an internucleotide bond, two nucleotides must be connected with the elimination of a water molecule. The resulting dimer then reacts with the next monomer (nucleotide) to form a trimer, and so on. For these reactions to take place, the following conditions must be met.
1. It is necessary to achieve the selectivity of the connection of monomer units. For example, the hydroxyl group of the first nucleotide should interact with the phosphate residue of the second, but not vice versa, because one of the principles of building polynucleotides in nature is unipolarity, i.e. monomer compound always in direction 5" → 3" . For this, fragments that do not participate in the reaction must be blocked using special protective groups.
2. By themselves, the hydroxyl and phosphate groups in nucleotides do not react with each other. To carry out the condensation of monomers, it is necessary to activate the phosphate group.
3. Since the synthesis is carried out in several stages, each reaction must proceed with a very high yield.
4. All processes - the introduction and removal of protective groups, activation and condensation - must be carried out under mild conditions, that is, without the use of high temperatures, as well as concentrated solutions of acids or alkalis, which could lead to the formation of by-products and the destruction of existing and newly formed chemical bonds.
At present, a large number of methods for the synthesis of oligonucleotides that satisfy these conditions have been developed, convenient protective groups have been selected, and methods for activating the phosphate group have been proposed.
Synthesis of extended fragments of nucleic acids consists of a large number of stages. After carrying out each chemical reaction, it is necessary to isolate the resulting product, purifying it from unreacted starting materials and other impurities. This makes the process long and laborious, and also leads to significant losses at each stage of isolation.
The American scientist Robert Merrifield in 1962 proposed the original idea of a solid-phase synthesis method, which made it possible to drastically simplify and speed up the process. The idea is that the first monomer (nucleoside) is attached to an insoluble polymer carrier (solid phase) which is placed in a reactor column. A solution containing the second monomer and other necessary reagents is passed through the column. In this case, the resulting reaction product also turns out to be attached to the solid phase. The column is then washed with solvent to remove unreacted substances and by-products, after which the next monomer is passed through the reactor, repeating the procedure many times until the synthesis of the desired product is completed. Thus, the growing polymer chain is fixed on the solid phase during synthesis, and all reactions with other components in solution proceed on the support surface. This technique makes it possible to replace complex and time-consuming procedures for separating and purifying intermediate products with elementary polymer washing operations.
The synthesis includes several fundamental stages.
Stage 1. Obtaining protected monomers, which are the starting blocks for building an oligonucleotide chain. To do this, those fragments of molecules that should not undergo chemical transformations are blocked with special protective groups.
Step 2: Attachment of the terminal monomer (protected nucleoside) to the polymer carrier. The carrier granules with attached terminal monomer are introduced into the reactor column.
Step 3: Deprotection of the terminal monomer. To do this, a solution of a reagent is passed through the column, causing the elimination of the protective group.
Stage 4. Condensation, for which a solution of the second monomer mixed with an activating reagent is passed through the column.
Steps 3 and 4 are repeated many times until a biopolymer of the required length is obtained. Then steps 5 and 6 are carried out.
Stage 5. Treatment of the support with reagents leading to the cleavage of the synthesized product from the solid phase and the removal of all protective groups.
Stage 6. Isolation and purification of the synthesized nucleic acid fragment using various chromatographic and electrophoretic methods. (Due to the incompleteness of the reactions, shorter chain fragments accumulate on the support towards the end of the synthesis, so thorough purification of the final product is necessary).
The main contribution to the development of oligonucleotide synthesis was made by G. Korana, who in the early 60s carried out the chemical synthesis of nucleic acid fragments of a given sequence and received the Nobel Prize for this work in 1968. In 1970, he first synthesized the complete alanine transfer RNA gene.
In 1975, R. Letsinger proposed a new method for the formation of an internucleotide bond, on the basis of which, in the early 80s, M. Carruthers developed the so-called solid-phase amidophosphite or phosphoramidite method for the synthesis of oligonucleotides. Since that time, the rapid development of this method and its automation began.
The initial building blocks for the synthesis of an oligonucleotide are modified deoxyribonucleosides (amidophosphites or phosphoramidites). These compounds containing trivalent phosphorus are more active than pentavalent phosphorus derivatives (Fig. 31).
Amidophosphite monomers are derived from protected nucleosides. The modification consists in attaching a benzene group to the amino groups of deoxyadenosine and deoxycidin, and an isobutyral group to the amino group of deoxyguanosine. Thymidine, which lacks an amino group, is not modified. This modification is necessary to protect the nucleosides from unwanted side effects.
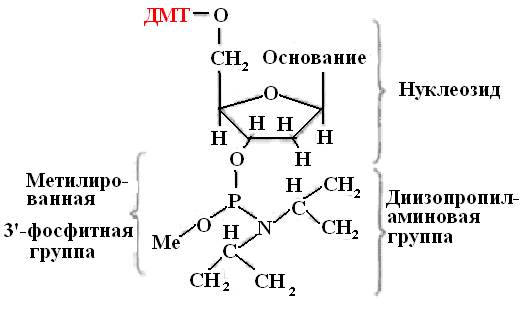
Rice. 31. Structural formula of phosphoramidite
Such derivatives of all four bases - A, T, G and C are used for the chemical synthesis of DNA.
DMT - dimethoxytrityl
Me - methyl group
Synthesis is carried out in the solid phase, the growing DNA chain is fixed on a solid carrier, which makes it possible to carry out all reactions in one container, easily wash out unnecessary reagents after each stage and add new ones in an amount optimal for the complete reaction. The first nucleoside is fixed to a solid support by chemical spacer(Figure 32). These are usually porous glass beads with pores of the same size.
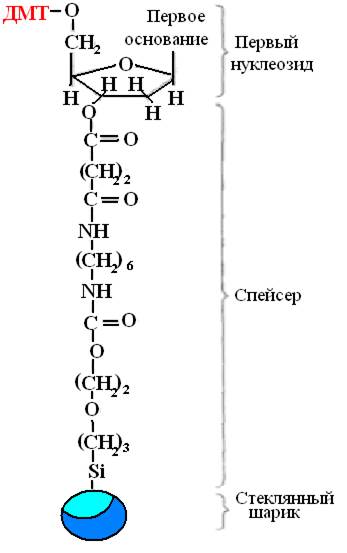
Rice. 32. The first nucleoside fixed with a spacer.
After n cycles carried out according to the synthesis scheme (Fig. 33), a single-stranded DNA fragment is formed from n + 1 nucleotide
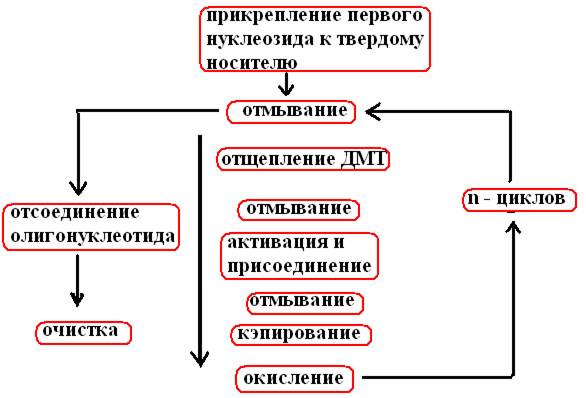
Rice. 33. Scheme of chemical synthesis of an oligonucleotide
The cycle begins after the attachment of the first nucleoside to the glass bead
Washed with an anhydrous solvent (acetonitrile), the remains of which are removed by purging with argon
Washed with TCA (trichloroacetic acid) to remove DMT (detritylation)
It is again washed with solvent and TCA, purged with argon, and at this stage the next nucleoside in the form of phosphorimidite and tetrazole, which activates phosphoramidite, are introduced, unbound reagents are removed by purging with argon.
Since, at the end of the first step, not all of the supported nucleosides are bound to the phosphoramidite, it is necessary to prevent their interaction with the nucleoside added in the second step. To do this, the unreacted 5'-OH group is acetylated with acetic anhydride and dimethylaminopyridine (capping).
The unstable phosphite triester bond formed in the second step between the nucleotides is oxidized with an iodine mixture to a pentavalent phosphate triester.
All the described operations are carried out until the last nucleoside is added to the growing chain in accordance with the program.
The methyl groups are removed by chemical treatment directly in the reaction column. Then, the oligonucleotides are detached from the spacer molecule together with the 3'-OH end and eluted from the column; then the benzoyl, isobutyral and DMT groups are sequentially removed. The 5'-end of the chain is phosphorylated by enzymatic (polynucleotide kinase + ATP) or chemical method
An important advantage of the solid-phase synthesis method is the simplicity of operations. The synthesis process consists of repeated steps of passing the protected monomers, reagents and solvents through a column with a polymer carrier to which a growing chain is attached. This makes it possible to automate the process.
Automatic synthesizers include a reactor containing a carrier with a first monomer attached to it, and vessels with the necessary monomers, reagents, and solvents. With the help of a pump, the solutions from these vessels are alternately fed into the reactor in accordance with the available program and the given sequence of the protein or nucleic acid fragment being synthesized. Currently, automatic synthesizers of peptides and oligonucleotides are produced by various companies, including domestic ones.
DNA sequencing
Comprehensive information about a DNA molecule can only be obtained by determining its nucleotide sequence. This procedure is called sequencing(from English. sequence). Thus, by sequencing the gene, i.e. having established the nucleotide sequence of a certain section of DNA, it is often possible to establish its function by comparing this nucleotide sequence with those for genes whose function is already known. Without data on the nucleotide sequence, it is impossible to conduct research on molecular cloning, as well as the creation of expression recombinant DNA.
DNA sequencing is quite expensive. They require special precision equipment, ultra-pure chemicals and biological preparations, as well as highly trained specialists. The potential of biotechnology will be fully realized only when its tools (including genome sequencing) become as accessible and inexpensive as personal computers. To reduce the cost of the sequencing procedure, the developers of new methods are trying to reduce the number of preparatory steps, miniaturize the equipment to the limit and sequencing millions of molecules simultaneously.
Before proceeding to the description of sequencing methods, it is necessary to recall the main points of reproduction of DNA nucleotide sequences, i.e. some fundamental steps of DNA replication.
The DNA of a cell starting to divide undergoes cardinal changes: the double helix unwinds, the chains diverge. On each chain, the synthesis of complementary polynucleotides begins, on one - continuous, on the second - discontinuous. It is catalyzed by an enzyme called DNA-dependent DNA polymerase; another enzyme, DNA ligase, links polynucleotide fragments into a continuous chain. So from one DNA molecule two are formed.
Many DNA sequencing methods are based on the mutual complementarity of the chains of this molecule. The genetic alphabet consists of only four letters - the nitrogenous bases of adenine (A), cytosine (C), guanine (G) and thymine (T). The bases of opposite strands of the DNA molecule are connected in accordance with the rule of complementarity: A forms a pair with T, and C with G. As a result of this interaction, the well-known double helix is formed - a structure resembling a spiral staircase (Fig. 34).
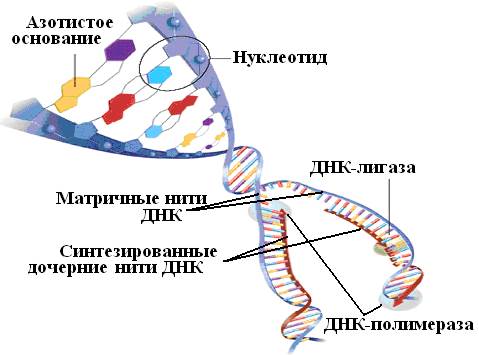
Rice. 34. Scheme for the synthesis of daughter nucleotide chains on both DNA chains
Living organisms use the principle of complementarity to copy their genetic material (replication) and repair damage to it (repair). It also underlies the amplification (see PCR) of target DNA fragments and their subsequent sequencing using a method developed in the late 1970s. F. Sanger.
